Freida Blostein was awarded $10,000 by ITiMS to conduct this project.
Co-mentors: Betsy Foxman and Kelly Bakulski
Testing the role of metabolic interactions and processes in the formation of cariogenic oral microbiomes
Early childhood caries (ECC), or dental decay in the primary teeth of children under 6 years of age, is a painful condition which negatively impacts self-esteem and predicts poor oral health throughout the life course (1–3). ECC affects 20% of children aged 2–5 in the United States (4). ECC is a multifactorial condition with social, behavioral, and microbial risk factors (5). The proximate mechanism involves microbial digestion of carbohydrates to enamel-eroding acids (5).
Streptococcus mutans was once considered the causal agent of ECC (5,6). S. mutans is strongly associated with ECC but is not always present in affected children or absent in healthy children. The ecological hypothesis of dental decay posits instead that bacterial communities interact, creating a functional shift to disease (5–7). Using metagenomics, our lab identified co-occurring microbial groups (guilds) which were prospectively associated with ECC and predicted S. mutans detection (8). ECC was also associated with the abundance of genes encoding metabolic and antibiotic functions. Our work suggests microbial interactions predict an ecological succession to cariogenic functions. Disrupting these interactions could prevent ECC.
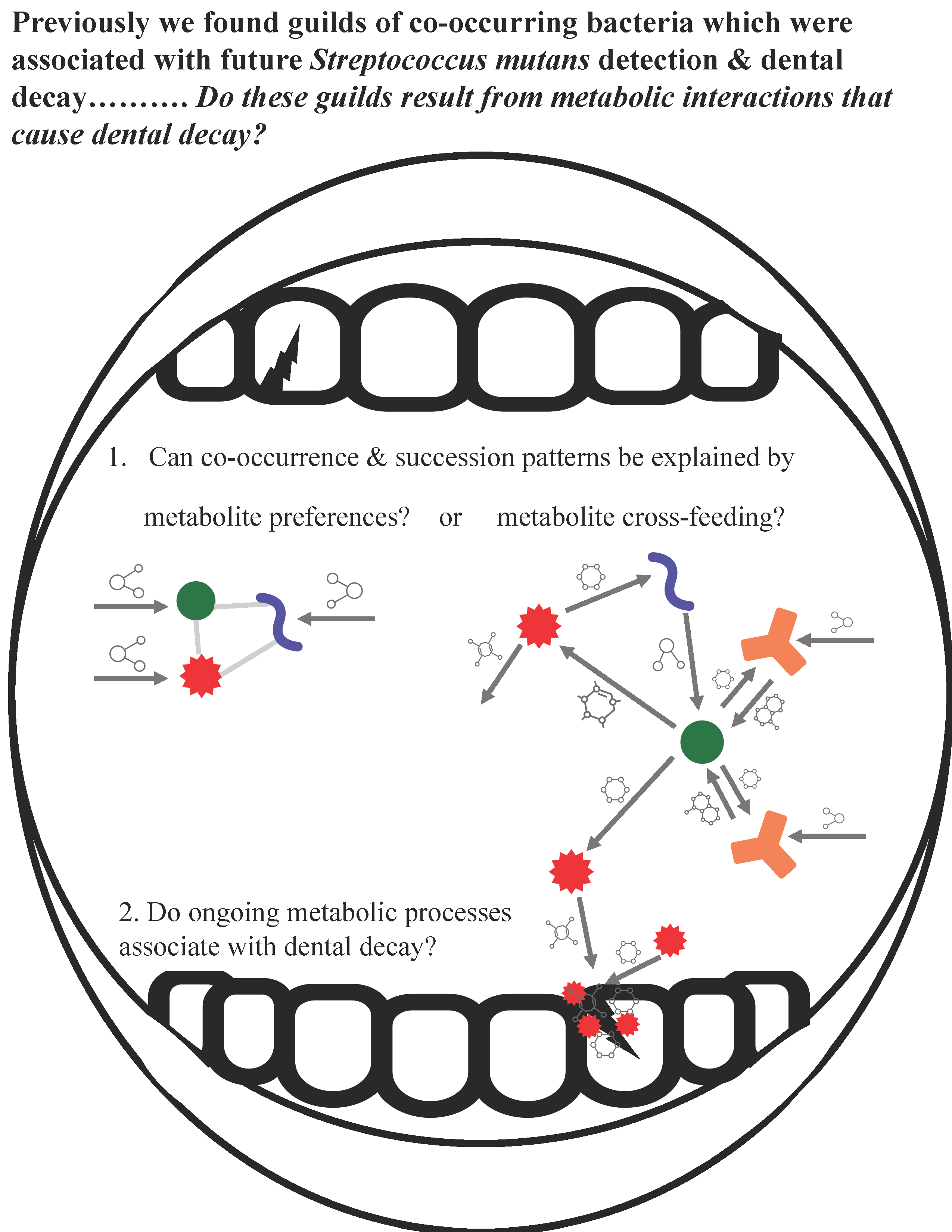
However, co-occurrence patterns observed in metagenomic data do not necessarily represent functional interactions. Observed correlations between microbes can result from habitat-filtering, in which organisms co-occur due to nutrient availability, rather than co-operation (9–11). Since microbial genes are not always expressed, metagenomic data does not measure ongoing functions but only their potential to be carried out. To address these gaps, I propose using agent-based models and RNA sequencing to test if metabolic interactions and processes explain assembly, succession, and cariogenicity of microbial communities.
Aim 1: Evaluate if previously observed co-occurrence and microbial succession patterns can be recapitulated in agent-based models of metabolic interactions.
Build in silico microbiomes using BacArena and publicly available genome-scale metabolic models.
a) Test if oral microbial co-occurrence guilds are selected for by a) initial nutrient availability or b) cross-feeding and cooperative growth.
Vary initial nutrient availability and community members independently and measure impacts on end community composition, metabolic exchanges, and biofilm growth.
b) Test if oral microbial co-occurrence guilds inhibit or promote S. mutans growth.
Cultivate S. mutans in different oral microbial communities and evaluate its biomass growth.
Aim 2: Evaluate if differences in ongoing metabolic processes associate with ECC and match agent-based model predictions.
Measure ongoing gene expression using RNA sequencing on already collected saliva from children with ECC and age-matched controls.
a) Test for differences in bacterial and host gene expression by early childhood caries status.
Compare gene expression of a) ECC cases vs controls at diagnosis b) ECC cases vs controls at the visit preceding diagnosis and c) ECC cases at diagnosis vs the preceding visit.
b) Compare in vivo species-specific gene expression with predicted metabolic fluxes from agent-based models.
Map gene data from expression data to metabolic reactions and phenotypes using Gene-Protein-Reaction rules and Boolean operators. Compare expression profiles in saliva samples to those in the agent-based model.
This project combines epidemiological data, agent-based models, and laboratory sequencing to better understand co-occurrence and succession in the early-life oral microbiome. Understanding if and how metabolic interactions between bacteria underly the formation of cariogenic oral communities can inform targeted interventions, such as prebiotics.
Text in graphical abstract:
Previously we found guilds of co-occurring bacteria which were associated with future Streptococcus mutans detection & dental decay………. Do these guilds result from metabolic interactions that cause dental decay?
1. Can co-occurrence & succession patterns be explained by metabolite preferences? or metabolite cross-feeding?
2. Do ongoing metabolic processes associate with dental decay?