Jinny Yang received a $5,000 award from ITiMS to conduct this project.
How do host-microbiome interactions shape the phytoplankton microbiome?
Introduction
Phytoplankton and their associated microbiomes of heterotrophic bacteria are foundational to primary production, energy transfer, and biogeochemical cycling in aquatic systems [1, 2]. While it is known that these microbiomes are influenced by host-released dissolved organic matter (DOM), the extent to which dynamic phytoplankton-bacteria interactions shape bacterial community assembly remains to be examined.To bridge the knowledge gaps, I investigated the effects of two mechanisms in host-microbiome interactions on phytoplankton bacterial microbiome formation: (i) innate host selection and (ii) host-microbiome feedback. For the former, phytoplankton-produced DOM composition is based solely on the host’s properties (species, genotype, or physiological state [3, 4]); for the latter, the presence and activity of the microbiome modify host DOM production [5–7]. Although both are often mentioned as important mechanisms in phytoplankton bacterial microbiome formation, we lack explicit tests examining the impact of each mechanism simultaneously.
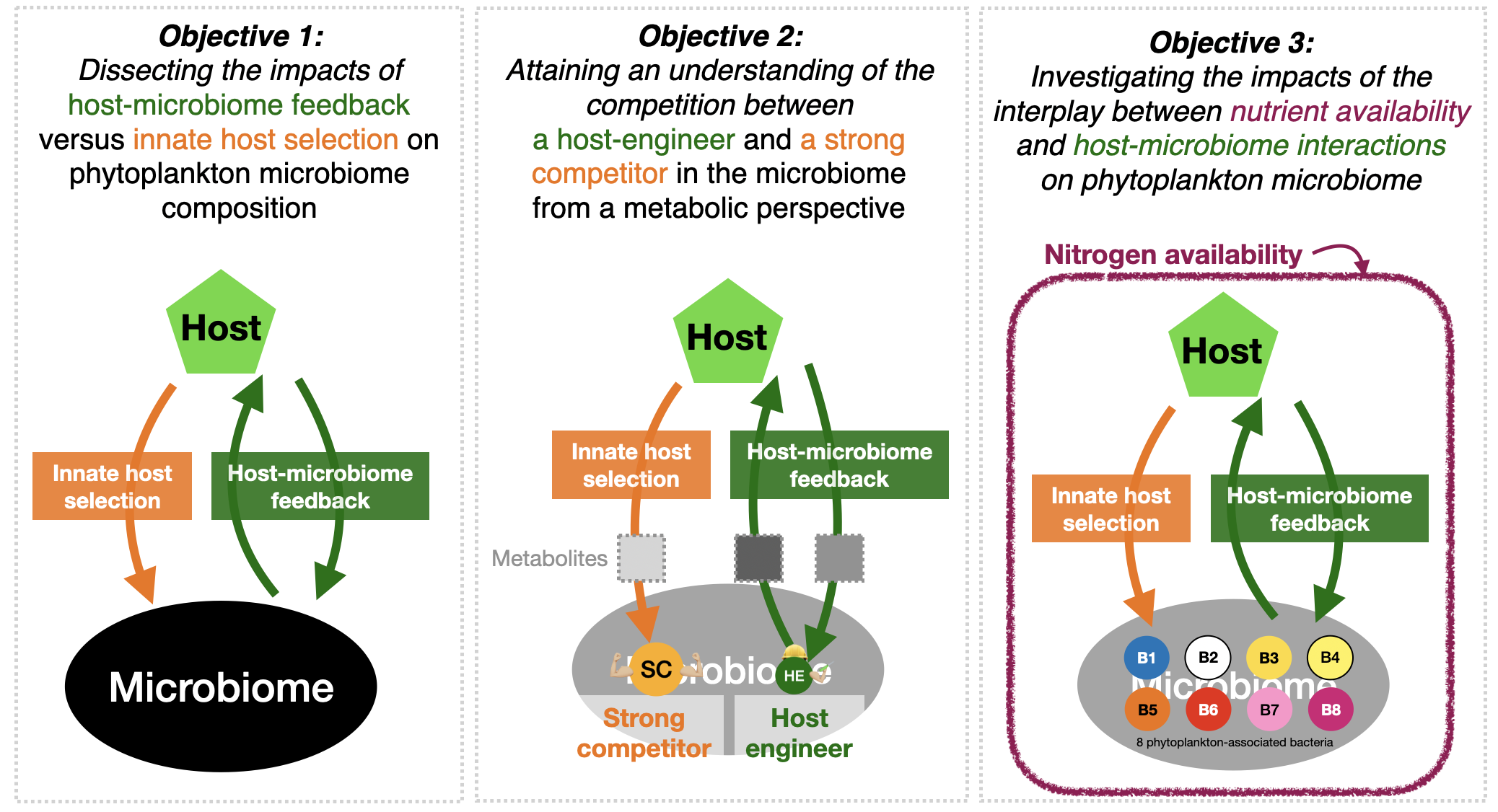
Objective 1: Dissecting the impacts of host-microbiome feedback versus innate host selection on phytoplankton microbiome composition
The first step of this project was to evaluate if the two mechanisms in host-microbiome interactions show a divergent or similar impact on microbiome assembly. The microbiome of Chlorella sorokiniana was extracted and exposed to six ratios of the two forces (hereafter feedback-to-innate index). We found that microbiome composition showed a continuous change along with the feedback-to-innate index, with the highest compositional distance between communities under the 0% and 100% feedback-to-innate indices (Figure 1). This indicates that each mechanism selects different bacterial species. In addition, the original phytoplankton microbiome was most similar to the microbiome when both mechanisms were applied, indicating that the natural phytoplankton microbiome is shaped by both forces rather than host-microbiome feedback or innate host control alone.
Objective 2: Attaining an understanding of the competition between a host engineer and a strong competitor in the microbiome from a metabolic perspective.
Knowing innate host selection and host-microbiome feedback showed a divergent impact on microbiome composition; here, I further explore the potential ecological processes that drive this divergent impact from the metabolic perspective. I hypothesize that the microbiome composition is the competition outcome between two functional groups of bacteria: (i) the host engineers (favored by host-microbiome feedback), who provide benefits to the host (e.g., growth hormones, vitamins, and siderophores) that change host’s metabolic status and the produced DOM composition, and in return, increase their fitness in the competition, and (ii) the strong competitors (favored by the innate host selection), who can consume the innate host DOM and uptake nutrients quickly but do not directly interact with the host. Therefore, I expect a shift in host metabolic status and the produced DOM composition when with the presence of host engineers, which would lead to increased fitness of the host engineer and its coexistence with the strong competitor in the microbiome.
To test the hypothesis, two bacterial representatives of a host engineer (favored by host-microbiome feedback) and a strong competitor (favored by innate host selection) were identified from a collection of bacteria isolates of C. sorokiniana microbiome. The two bacteria were grown in mono-culture and mixed-culture under innate host selection and host-microbiome feedback effect. Control treatments without bacteria were also conducted. After incubation, cultures were collected for transcriptomic and metabolomic analysis to reveal the metabolic transcriptional profile in bacteria and phytoplankton host and the composition of extracellular metabolites in the culture medium.
Objective 3: Investigating the impacts of the interplay between nutrient availability and host-microbiome interactions on phytoplankton microbiome.
Finally, I increase the microbiome diversity to 8 bacterial species and evaluate how bacterial functionality in host-microbiome interactions (strong competitor versus host engineer) affects their competition outcome in the microbiome. In addition, I consider the impact of environmental nutrient availability which is known to influence phytoplankton-bacteria interaction morphology [8–10]. The growth curve of 8 phytoplankton-associated bacteria under host-microbiome feedback and innate host selection in function of three different nitrogen concentrations (which represented the oligotrophic, eutrophic, and polluted lake system) will be measured. These growth parameters will be used to develop mathematical modeling to predict how microbiome composition shifted with nutrient availability under the host-microbiome interactions, which will be tested by another culture experiment on 8-bacterial isolates mixed.
Reference:
1. Alvarenga DO, Rousk K. Unraveling host–microbe interactions and ecosystem functions in moss–bacteria symbioses. Journal of Experimental Botany 2022; 73: 4473–4486.
2. Wilkins LGE, Leray M, Yuen B, Peixoto R, Pereira TJ, Bik HM, et al. Host-associated microbiomes and their roles in marine ecosystem functions. 2019. PeerJ Preprints.
3. Tada Y, Nakaya R, Goto S, Yamashita Y, Suzuki K. Distinct bacterial community and diversity shifts after phytoplankton-derived dissolved organic matter addition in a coastal environment. Journal of Experimental Marine Biology and Ecology 2017; 495: 119–128.
4. Zhou J, Richlen ML, Sehein TR, Kulis DM, Anderson DM, Cai Z. Microbial community structure and associations during a marine dinoflagellate bloom. Frontiers in Microbiology 2018; 9: 1–21.
5. Mühlenbruch M, Grossart H, Eigemann F, Voss M. Mini‐review: Phytoplankton‐derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environmental Microbiology 2018; 20: 2671–2685.
6. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of phaeobacter gallaeciensis. Nature Chemistry 2011; 3: 331–335.
7. Shibl AA, Isaac A, Ochsenkühn MA, Cárdenas A, Fei C, Behringer G, et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proceedings of the National Academy of Sciences 2020; 117: 27445–27455.
8. Mayali X. Editorial: Metabolic interactions between bacteria and phytoplankton. Frontiers in Microbiology 2018; 9: 1–4.
9. Diner RE, Schwenck SM, McCrow JP, Zheng H, Allen AE. Genetic Manipulation of Competition for Nitrate between Heterotrophic Bacteria and Diatoms. Front Microbiol 2016; 7.
10. Le Chevanton M, Garnier M, Lukomska E, Schreiber N, Cadoret J-P, Saint-Jean B, et al. Effects of Nitrogen Limitation on Dunaliella sp.–Alteromonas sp. Interactions: From Mutualistic to Competitive Relationships. Front Mar Sci 2016; 3.
Figure 1. Principal Coordinates Analysis (PCoA) ordination based on Bray-Curtis dissimilarities between phytoplankton microbiome communities in the function of feedback-to-innate index at the final semi-incubation day (after five times bi-daily transfer). Symbols represent the microbiome communities under 0% (open circle), 5% (open triangles), 25% (gray circles), 50% (gray triangles), 75% (black circles), and 100% feedback-to-innate index (black triangles), respectively. Star symbols represent the original bacterial microbiome communities on the initial day (microbiome prior to detachment procedure from C. sorokiniana).